 username@email.com
username@email.com
In this lesson, you will review the process by which DNA and RNA are replicated in a cell.
The hydrogen bonds holding the base pairs in a DNA molecule together are relatively weak compared to other chemical bonds. Even hot water is enough to cause the base pairs to separate. Watson and Crick reasoned that, since the hydrogen bonds were weak, the double helix of DNA somehow “unzipped” into two strands. So DNA replication would occur in a few simple steps:

The Watson and Crick model for DNA replication is also known as a semi-conservative model. Two other models of DNA replication were considered. In the conservative model, the parent DNA remains intact and an all new copy is created. In the dispersive model, each strand of both copies contains a mixture of old and new DNA fragments.
Before biologists could tackle the problem of the details of DNA replication, the correct model had to be identified. Matthew Meselson and Franklin Stahl devised an experiment using different isotopes of nitrogen in bacterial cultures. First, several generations of E. coli bacteria were cultured in a medium that contained a heavy isotope of nitrogen, 15N. These bacteria were then transferred to a medium containing only the lighter isotope, 14N, and cultured for one more generation. It was hypothesized that any new DNA synthesized during this single replication would be less dense than the original DNA because it would have an all 15N DNA strand and an all 14N DNA strand. This difference in density would show up when the extracted DNA was centrifuged. This is exactly what was found and this result eliminated the conservative model from consideration. However, the dispersive model would also produce DNA with an intermediate density. In order to decide which of the remaining two models was correct, the remaining bacteria were allowed to replicate a second time. According to the dispersive model, the resulting DNA would still have a range of intermediate densities.
According to the semi-conservative model, two different densities of DNA would be present. One would contain DNA with an all 15N DNA strand and an all 14N DNA strand. However, an even less dense DNA would also be present that had two strands containing only newly synthesized all 14N strands. The results showed DNA of only two different densities, exactly as predicted by the semi-conservative model. The diagram below shows the results.

In double-stranded DNA, how do the four bases (C, A, T, G) pair up with each other?
C is the correct answer. Choices A and D are incorrect because bases do not pair with themselves. B is incorrect because due to size restrictions, purines always pair with pyrimidines.
Which of the following is the name of the process through which DNA replicates?
B is the correct answer. The experiments of Meselson and Stahl showed that second generation DNA was a combination of original strands with new strands and third generation DNA also contained some completely new chromosomes. Conservative replication would preserve the original chromosome. Dispersive replication would have produced all intermediate chromosomes with a range of densities. While it might seem logical that there is a liberal model to contrast with the conservative model, no such model exists.
Watson and Crick’s model of semi-conservative replication was supported by experimental evidence, but there were many details yet to be worked out. What caused the strands to separate? How did new bases find their way to the correct spots on the template? What caused the nucleotide bases to connect to form the backbone? What provides the energy for all this to happen?
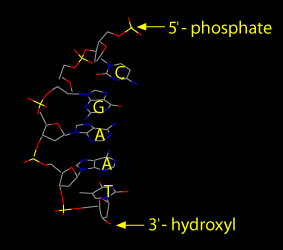
Each DNA strand has a uniquely identifiable directionality. The two ends of a single DNA strand are different. The five carbon atoms of one deoxyribose sugar of each DNA strand are numbered, from 1′ to 5′ (‘ represents prime). A nucleotide’s phosphate group is attached to the 5′ carbon atom of deoxyribose. The phosphate group of one nucleotide is joined to the 3′ carbon atom of the adjacent nucleotide. Thus the DNA strand has a definite polarity. At one end, called the 5′ end, a phosphate group attached to the 5′ atom of the last nucleotide terminates the strand. At the other end, called the 3′ end, a hydroxyl group attaches to the 3’ atom of the terminal deoxyripose.

The energy that drives this whole process comes from the nucleotides. The nucleotides that serve as substrates for DNA polymerase are nucleotide triphosphates. This molecule is very similar to ATP. The only difference is the sugar component, deoxyribose instead of ribose. As each monomer joins the growing end of a DNA strand or Okazaki fragment, it loses two phosphate groups. Hydrolysis of these phosphates provides the energy.
Which of the following enzymes is not involved in DNA replication?
C is the correct answer. Phosphatase is an enzyme that releases phosphates for cell metabolism. It is not involved in DNA replication. The other three enzymes play central and critical roles in DNA replication.
What is the name given to the short fragments of DNA produced on the lagging strand?
B is the correct answer. Suzuki is the name of a method for teaching music, a brand of motorcycle, and a brand of automobile. Osaka and Hirosaki are the names of Japanese cities and universities.
Prokaryotes, such as bacteria, have a single circular chromosome containing a closed-loop DNA molecule that contains about five million base pairs. Replication of this molecule starts at a single point and proceeds in both directions until complete. In E. coli, the rate of replication is about 1,000 base pairs per second, so replication is complete in about one hour. Human cells contain 46 chromosomes, each containing a DNA molecule with about 150 million base pairs. The copying rate in human cells is only about 50 base pairs per second. So, if human DNA replicated the same way as prokaryotic DNA replicates, it would take over one month to complete one copy.
Fortunately, there are many places on the eukaryotic chromosome where replication can begin. Replication begins at several “replication bubbles” and proceeds in both directions from each bubble.
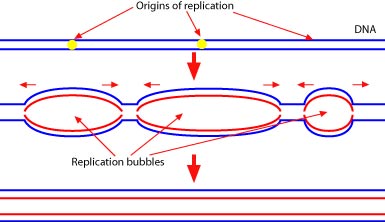
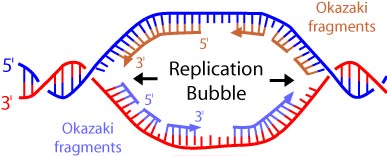
Which of the following is an important difference between prokaryotic and eukaryotic replication?
A is the correct answer. Since eukaryotic chromosomes contain many more base pairs, replication must occur at several different sites simultaneously. Choice B is incorrect because the rate of prokaryotic base pairing is much faster. Choice C is not a difference, both processes proceed in both directions. Choice D is not a difference since both prokaryotic and eukaryotic replication are mediated by several different enzymes.
The accuracy of DNA replication is not due solely to the specificity of base pairing. Errors in the completed DNA molecule probably occur in only one out of one billion nucleotides, but initial errors do occur about one in 10,000 base pairs. These mismatches are detected by DNA polymerase during replication. Other proteins also perform mismatch repair. Failure to repair mismatches can lead to accumulated genetic defects.
DNA molecules in a cell are subjected to a variety of chemical and physical assaults, such as damage by ultraviolet, x-rays or chemicals. Although most mutations are not harmful, left unrepaired, defects in DNA can accumulate and may lead to a disorder or certain forms of cancer. Maintenance of DNA in a cell is so important that hundreds of different DNA-repair enzymes have evolved. Most work by some form of excision. The damaged area of the DNA is excised by a DNA-cutting enzyme, such as nuclease, and the gap is then filled in by DNA polymerase and ligase.