 username@email.com
username@email.com
In this lesson, you will review how the genetic information contained in DNA determines the inherited traits of an organism, and how genetic mutations occur.
The idea that genes dictate phenotypes through enzymes that catalyze specific chemical processes in the cell was first proposed in 1909 by the British physician Archibald Garrod. However, it was several decades later that experiments were done to test this idea. George Beadle and Edward Tatum studied mutations in bread mold, Neurospora crassa. The wild-type bread mold could exist with a minimal growth medium, such as agar. The mold synthesized everything else it needed. Beadle and Tatum identified three strains of Neurospora crassa, called auxotrophs, that required additional nutrients (e.g., amino acids, vitamins, and other nutrients) to survive and required a complete growth medium containing the additional nutrients. To determine a particular strain’s genetic defect, Beadle and Tatum took samples of the mutated strains and grew them in several different vials, each containing minimal medium plus a single additional amino acid.

Wild-type Neurospora crassa requires only minimal nutrients to survive. The mold uses multiple steps to synthesize several different essential amino acids, which are the building blocks of proteins. One process synthesizes arginine in three steps from a precursor, each step mediated by a specific enzyme. Each mutant class had a missing or defective gene which caused a block (marked by an X) in the synthesis process.
Further experiments have slightly modified the one gene/one enzyme hypothesis. Since many proteins, such as keratin, are not enzymes, biologists first suggested a one gene/one protein model. Currently, the most useful model seems to be one gene-one polypeptide. However, most biologists still call it the one gene/one protein model.
Which of the following describes a gene?
B is the correct answer, according to the one gene/one enzyme model. Choice A is incorrect because chromosomes contain thousands of genes. Choice C is a description of a codon. Choice D is a description of messenger RNA (mRNA).
Beadle and Tatum’s experiment demonstrated that each kind of mutant bread mold lacked a specific enzyme. This was conclusive evidence that…
The correct answer is A. Genes contain the information for making specific proteins. Beadle and Tatum did use radiation to induce the mutations, but this was known before the experiment, therefore choice B is incorrect. C is also not correct; although enzymes can repair DNA, Beadle and Tatum’s experiment was not proof. While it is true that cells need specific enzymes, this was the basis of the experiment, not the result, therefore choice D is incorrect.
Genes do not build proteins directly. The intermediary is RNA, a molecule similar to DNA, but RNA has ribose instead of deoxyribose as its sugar backbone. RNA also contains the nitrogenous base uracil instead of the chemically similar thymine. Furthermore, RNA almost always occurs as single strands instead of the double stranded helix of DNA.
The simplified schematic pathway of poltpeptide synthesis is:
DNA → mRNA → tRNA → rRNA (ribosome) → protein
There are slight differences in this process between prokaryotes and eukaryotes. Since prokaryotes lack a nuclear membrane, the mRNA moves directly from the relatively simple DNA molecule to the ribosome. In eukaryotic cells, precursor RNA is created first and the cell’s nuclear machinery then modifies this into mRNA, which then moves through the nuclear membrane and to the cytoplasm.
What is the name of the process by which the information contained in a DNA molecule is copied onto a messenger RNA molecule?
A is the correct answer. The DNA sequence is transcribed onto mRNA. Choice B is the name of the process where the mRNA information is used to construct polypeptides. Choice C is incorrect because the strand is not copied exactly, so it is not replicated. Choice D is not a term used in cell processes.
There are 20 different amino acids, which are the building blocks of all proteins, but there are only four different nucleotide bases. So, each amino acid must be coded by a sequence of nucleotide bases. Three bases (4 x 4 x 4 =64) produce 64 different combinations—more than enough to code for the 20 amino bases. So the fundamental “letter” of the genetic “alphabet” is a sequence of three nucleotide bases, read in the 5′ to 3′ direction. These triplets are called codons.
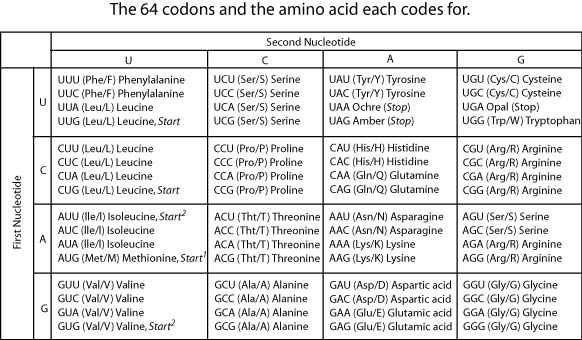
Two nucleotide bases would not be sufficient to code for the amino acids because two bases would sequence only
C is the correct answer. Choices A, B, and D are all incorrect because the number of possible combinations of four letters taken two at a time is 4 x 4 = 16.
The RNA codon CCG is translated as the amino acid proline in all organisms whose genetic code has been examined. The only differences in this commonality of the genetic code to all organisms occur in some very simple eukaryotes, such as Paramecium. The DNA of some specialized cell organelles, such as mitochondria and chloroplasts, also show minor differences.
A consequence of the universality of the genetic code is that pieces of DNA can be clipped out of one organism and inserted into another. Most of the world’s insulin supply is produced by bacteria that have been genetically modified in just this way.
Before the DNA code can be transcribed, the two strands of DNA must be separated by the enzyme, RNA polymerase which chemically “unzips” the strands. The process begins at a codon identified as a promoter on the DNA molecule and ends at a codon called the terminator. The transcription unit between promoter and terminator is a gene. After binding to the promoter, the RNA polymerase begins moving from the 3′ end to the 5′ prime end of the DNA strand, unwinding and separating as it moves. Nucleotide bases are matched to their pairs by the RNA polymerase. After the RNA polymerase reaches the terminator, the RNA strand is released. While the RNA polymerase moves in the 3′ to 5′ direction, because of the pairing rules, the RNA strand is assembled from the 5′ end to 3′ end.
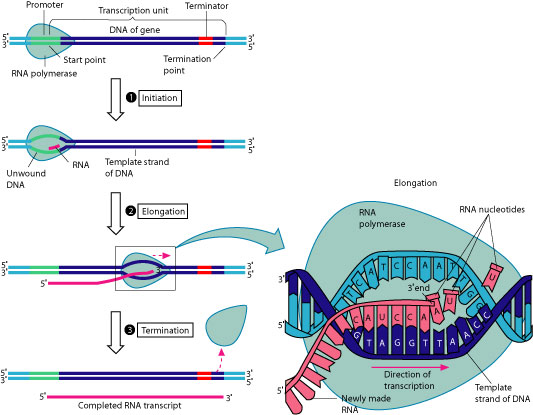
In a cell, the nucleotides GAT were temporarily paired with the nucleotides CUA. Where did this pairing most likely occur?
C is the best answer because in RNA, uracil replaces thymine. Uracil rarely occurs anywhere else but in RNA, therefore choice A is incorrect. Choice B is incorrect because nucleotide bases are not paired during translation. Choice D is incorrect because RNA is single-stranded.
In prokaryotes, RNA is immediately released into the cytoplasm where it is available to build proteins in the ribosomes. However, in eukaryotes, RNA is transformed in two different ways before it is released from the nucleus. Each end of the pre-mRNA molecule is modified by the addition of a “cap.” At the 5′ end, a guanine nucleotide is added. At the 3′ end, a “tail” is added, consisting of many adenine nucleotides. Both caps and tails serve to protect the ends of the molecule from degradation by other enzymes that may be floating around. The caps also serve as signals, telling the ribosome where to attach.
Another modification to mRNA is the removal of non-coding segments called introns. Surprisingly, the polypeptide codes are not continuous. The non-coding parts must be removed, leaving behind the parts that will code for proteins, called exons. This RNA splicing is carried out by small nuclear ribonucleosomes or snRNPs. (Some biologists call them “snurps.”) The snRNPs recognize splice sites and together with other proteins they form a larger enzyme-like assembly called spliceosome. The spliceosome cuts out the introns and splices the ends of the RNA back together.
After a precursor mRNA molecule is transcribed from a eukaryotic gene, it is modified. ____ are cut out and ____ are spliced back together to produce a molecule of mRNA.
D is the correct answer. The introns are not expressed in the protein, so they are removed and the exons are spliced back together. Choices A and B are incorrect because terminators and promoters occur on the DNA molecule and mark genes or transcription segments. The terms are reversed in choice C and are therefore incorrect.
It may seem like a waste of good nucleotides to have all of the introns mixed up in the genetic code. However, biologists think they may serve very important roles. It is known that the same gene can code for different proteins, depending on which parts are treated as introns. This is more efficient than having a different gene for every protein. Another role may be in the evolution of new and potentially useful proteins. Exon shuffling and gene recombination might create new, novel, useful, and important proteins.
Translation of mRNA to proteins takes place in a cell structure known as a ribosome. These structures allow specialized molecules known as transfer RNA (tRNA) to transfer amino acids from the cell’s cytoplasm to the ribosome. The ribosome attaches the amino acid brought to it by the tRNA to the growing polypeptide chain, according to the codes on the mRNA strand. The cell keeps its cytoplasm stocked with all 20 amino acids, either by absorbing them from nutrients outside the cell or by synthesizing them from other compounds.

The whole cycle of transcription and translation can happen simultaneously in prokaryotes. Translation of the mRNA to a polypeptide occurs as soon as one end of the strand of RNA is available. The mRNA may start moving through a ribosome while the other end is still being synthesized. Since the mRNA remains intact during the whole process and acts only as a template, a single strand of mRNA will eventually loop through many different ribosomes, all of which are busily producing polypeptides at the same time.
The molecules of tRNA are specialized. Each attaches to a single amino acid. The other end of the tRNA contains an anticodon, a nucleotide sequence complementary to the sequence on the RNA. This attaches to the RNA at the appropriate site according to the base pairing rules and is carried into the ribosome where the amino acid on the other end is attached to the lengthening polypeptide chain. The tRNA then detaches from the RNA as it slides out of the ribosome. It then is carried away into the cytoplasm where it attaches to another amino acid molecule. So the tRNA is continually recycled and thus acts as a catalyst to the reaction.

A tRNA molecule is only about 74–93 nucleotides long. This short strand of RNA folds back on itself and hydrogen bonds form between complementary base pairs. The overall shape of the tRNA is like an “L.” However, it is commonly represented flattened out into a structure resembling a four-leaf clover.
The machinery for attaching the proper amino acid to the appropriate tRNA molecule is provided by a specific enzyme called an aminoacyl-tRNA synthetase. There are 20 of these enzymes in the cell, one for each amino acid. The reaction binding the amino acid to tRNA is endergonic, so the enzyme uses energy from ATP to complete the process. After the amino acid is joined to tRNA, the enzyme is free to catalyze another reaction.
Transfer RNA is produced by the same genetic machinery as mRNA. The genes that produce tRNA are found on all chromosomes, except chromosome 22 and the Y chromosome. There are also genes in human cells that code for ribosomal RNA (rRNA) and there are genes found in the cell mitochondria that code for other tRNA.
An mRNA molecule with a complementary codon is transcribed from the DNA codon ACT. In the process of protein synthesis, a tRNA pairs with the mRNA codon. What is the nucleotide sequence of the tRNA anticodon?
C is the correct answer. The anticodon on the tRNA will match the original codon on the DNA except that uracil will replace thymine. The tRNA anticodon will pair up with a codon on the mRNA molecule following base pairing rules. Choice A is incorrect because this is the sequence on the mRNA molecule. Choice B is incorrect because it contains the base thymine. In RNA molecules, thymine is replaced by uracil. Choice D has the correct bases but in reversed sequence.
In prokaryotes, the RNA is immediately released into the cytoplasm where it is available to build proteins in the ribosomes. However, in eukaryotes, the RNA is transformed before it is released from the nucleus. So prokaryote mRNA is immediately available to begin translation. In eukaryotes, the mRNA must first be released from the nucleus.
Mendel thought of a gene as a discrete inheritable unit that produces some phenotypic character. Morgan and others identified genes as specific locations on a chromosome. Then a gene was identified as a specific nucleotide sequence along a molecule of DNA. Finally, a gene is functionally defined as a specific nucleotide sequence that codes for a specific polypeptide. This one gene/one enzyme model is still not completely satisfactory, because eukaryotic genes contain non-coding segments (introns). Different biologists also use somewhat different definitions, such as whether the promoter and terminator segments are included as part of the gene. The definition of a gene must also include the genes that code for rRNA, tRNA, and other types of RNA that do not code for polypeptides.
Mutations are changes in the genetic material of a cell or virus. Large-scale mutations can occur when a cell’s chromosome structure or number is altered in some way. Point mutations are changes in one or a few base pairs in a single gene.
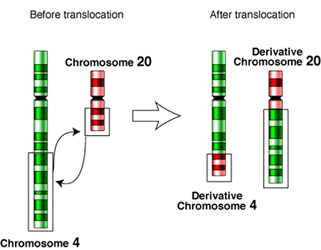
Changes in the number or structure of human chromosomes can produce phenotypical characters that are unlike the parents and that may be harmful. Alterations of the number of each type of chromosome are known to produce several different genetic disorders. Aneuploidy can produce cells that are monosomic, meaning they contain only one copy of a particular chromosome. Anueploid cells can also be trisomic, meaning they contain three chromosomes of a particular type. Down syndrome is caused by cells containing three copies of chromosome 21. Klinefelter syndrome (occurs in males: adults are mentally normal, have male sex organs, and develop breasts and a typical female body conformation during adolescence) is caused by possessing two X chromosomes and one Y Translocationchromosome. On the other hand, males with XYY and females with XXX both appear perfectly normal and are indistinguishable from XX or XY counterparts except by karotype.
Alterations of chromosome structure can result from deletions, duplications, inversions, or translocations. Deletions occur when sections of the chromosome are left out during cell division. Sometimes the missing fragment will become part of the homologous chromosome, producing duplication. If the fragment reattaches in the opposite direction, it causes a chromosomal inversion. Finally, translocations occur when two non-homologous chromosomes exchange fragments.
How many individual chromosomes (not pairs) does each cell in an individual with Down syndrome have?
D is the correct answer. People with Down syndrome have an extra copy of chromosome 21, for a total of 47 chromosomes. Choices A and C are both incorrect because humans normally have 23 pairs of chromosomes or 46 individual chromosomes. Choice B refers to chimpanzees, which have 24 pairs of chromosomes.
Point mutations are changes in one or a few base pairs in a single gene. These can be substitutions, insertions, or deletions. Insertions and deletions result in frameshifts. Substitutions occur when the wrong base is inserted into a gene during replication. The mistakes are usually repaired by the cell machinery, but about once in one billion the final copy of the DNA has a mistake.
Harmful substitutions can be missense or nonsense substitutions. Missense substitutions result in the production of an incorrect amino acid in a protein, thus changing the protein’s function. Nonsense substitutions change a codon into a STOP codon, prematurely terminating a protein. The shorter the protein is, the less likely it will function as it should.
One wrong base in a sequence might have no effect on the organism at all. The mistake could occur in an intron, so protein production is not affected. In other cases, the wrong base might be in the third position in a codon. There are many synonymous codons, so the wrong base might not result in the wrong amino acid in the polypeptide. These are called silent mutations, because they have no effect.

This figure shows how by shifting the reading frame one nucleotide to the right, the same sequence of nucleotides encodes a completely different sequence of amino acids.
Insertions and deletions are more likely to cause problems because either can result in a frameshift. Unless an insertion is extremely long, extra base pairs in an intron will have no effect. One exception is the Fragile X syndrome. This defect occurs when hundreds of CGG sequences are written into the human X chromosome. This causes the X chromosome to bunch up or fold over on itself or to be restricted in some other way. Males who inherit such a chromosome show a number of harmful phenotypic effects including mental retardation. Females who inherit a fragile X are only mildly affected, if at all.
When a frameshift occurs in a gene sequence that will be expressed as a protein, an insertion of three bases will cause one extra amino acid to be inserted in the protein. This may be relatively benign. However, the insertion or deletion of one or two base pairs will cause extensive changes in the genetic code. All of the nucleotides “downstream” from the insertion will be incorrectly grouped into codons. Unless the frameshift occurs very near the end of the gene, it is likely to result in a nonfunctional protein.
Which statement could explain why a particular point mutation had no affect on the polypeptide expressed by a gene?
D is the correct answer. If the substitution occurred in the third place in a nucleotide triplet, the expressed amino acid might not be changed. Choices A and C are unlikely because a deletion or insertion would cause a frameshift. Choice B is unlikely because the stop codon would prematurely terminate the polypeptide chain, almost certainly making it nonfunctional.
In any population of organisms, a range of phenotypes is usually present. This variation is vital to the survival of a population. The broader the range of expressed phenotypes, the more likely it is that some members of the population will survive sudden environmental catastrophes or slow climate changes. There are two primary sources for this variation. One is any mutation within a gene. The other is the reshuffling and recombination of chromosomes during sexual reproduction.
Point mutations are changes in one or a few base pairs in a single gene. These can be substitutions, insertions, or deletions. Substitutions occur when the wrong base is inserted into a gene during replication. The mistakes are usually repaired by the cell machinery but in about one time in one billion, the final copy of the DNA has a mistake.
Substitutions often cause no harm, but insertions and deletions result in frameshifts. When a frameshift occurs in a gene sequence that will be expressed as a protein, all of the nucleotides “downstream” from the insertion may be incorrectly grouped into codons. Unless the frameshift occurs very near the end of the gene, it is likely to result in a nonfunctional protein.
Among species that reproduce sexually, the chromosomes in each generation are shuffled and mixed in a variety of possible combinations. During meiosis, each of the two homologous chromosomes are separated and sorted into gametes independently of the others. Each individual received a set of chromosomes from each parent of that individual. A gamete produced by that individual could conceivably contain only the chromosomes from one parent, but is far more likely to receive a random mixture of chromosomes from each parent. Since humans have 23 chromosome pairs, there are 223 possible combinations of chromosomes in each gamete—that is around 8 million possible combinations! Since an offspring receives a set of chromosomes from each parent, each set having 223 possible combinations of chromosomes from the grandparents, it is obvious that there is an enormous number of combinations of chromosomes in each generation, assuring a wide range of phenotypes.

Since chromosomes are randomly mixed but not destroyed during sexual reproduction, the same set of genes, commonly called the gene pool, is preserved for future generations unless something happens to the population. A large gene pool is most desirable. Small gene pools mean that a disease or some other environmental problem could wipe out an entire population. For example, cheetahs the world over are genetically almost identical. Biologists speculate that something wiped out most of the world’s cheetahs, leaving only a small population with limited genetic diversity to repopulate the cheetahs.
Which of the following processes is a result of environmental pressure on the genotypic variation in populations?
D is the correct answer. Evolution requires changes in the genotypes of a population over time. Choices A, B, and C are all incorrect because cell division does not require variation.
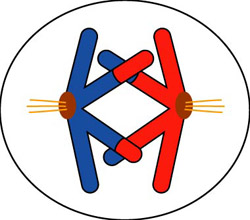
Sometimes, chromosomes will “cross over” during meiosis. During Prophase I, the homologous chromosomes adhere closely together, something like the halves of a zipper. The pairing is precisely gene-by-gene. During this phase, the homologous chromosomes can exchange segments. So, in addition to the many possible combinations of chromosomes from each parent, the crossing over process produces new and novel but completely viable chromosomes. This is an important source of genetic variation that results from sexual reproduction.
Which of the following statements describes crossing over?
A is the correct answer. When homologous chromosomes exchange corresponding genes, novel but viable chromosomes are created. Choice B is an important process that produces new combinations of chromosomes but does not change the individual chromosome. Choice C is the process of translocation, which typically produces nonviable or harmful changes in chromosome structure. Choice D is a description of errors that occur during gene replication, not meiosis.